Library
- PharmaLogical® Library
PharmaLogical® Library is EME’s humanized synthetic VHH library was constructed in collaboration with Mitsui Knowledge Industry Co., Ltd., (HQ: Tokyo, Japan; President & CEO: Kengo Asano; “MKI”).
- The concept of PharmaLogical® Library
Structure based
DesignMinimized
HeterogeneityLibrary Diversity
≧10¹³⁻¹⁴Humanized FRs
- Structure based Design ― Designed VHH antibodies with structural data of VHHs ―
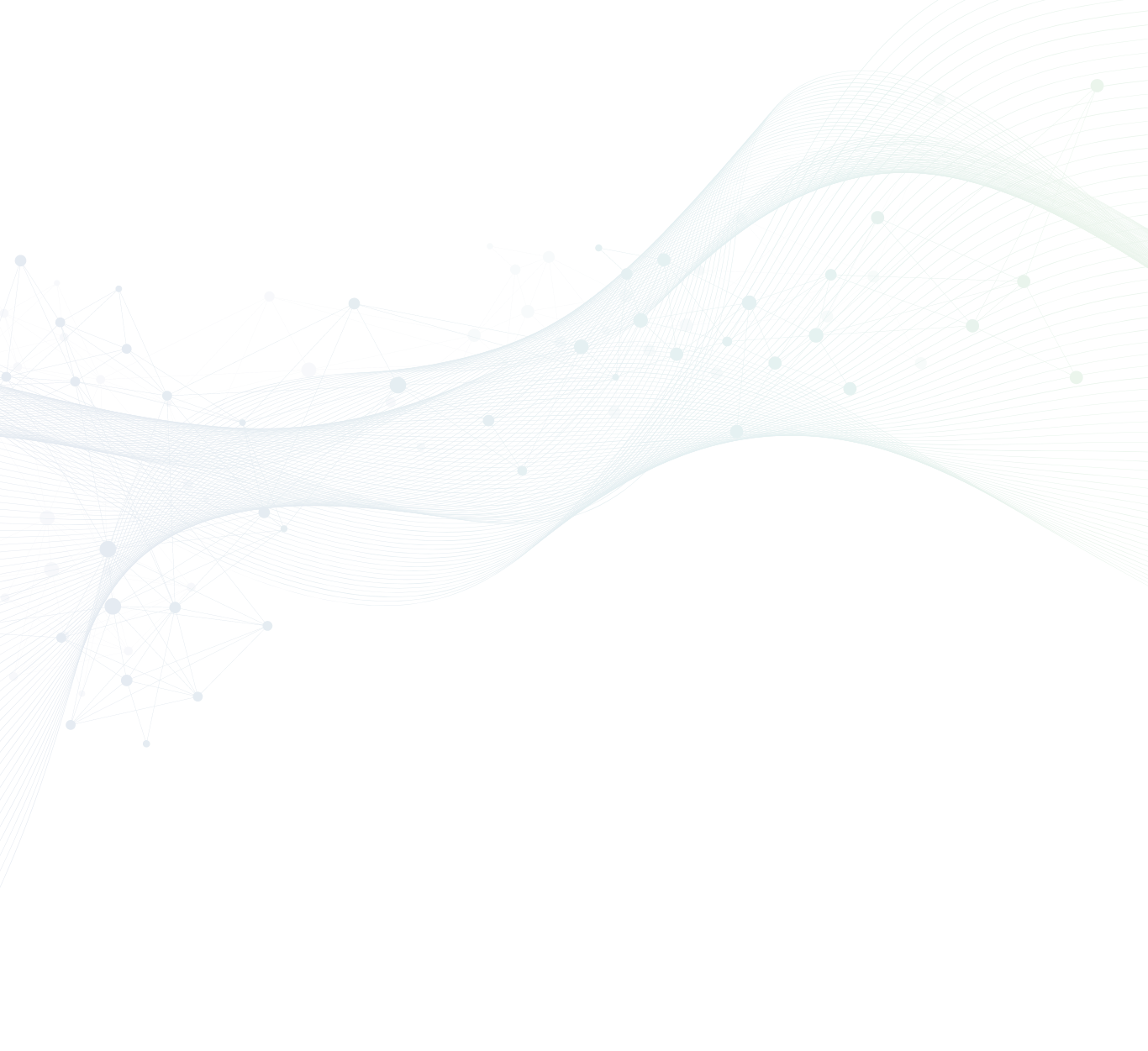
In collaborative research with Mitsui Knowledge Industry Co., Ltd., we analyzed more than 400 crystal structure analysis data of camelid VHHs. From the data analysis, we successfully classified VHHs by CDR3 loop structures which are important for binding to targets. The structures of VHHs are classified into three types based on the conformation of CDR3; (A)Upright, (B)Half-Roll and (C)Roll (PDB ID: 5j1s, 6ru3, and 2p45). Also, we found that there is a relationship between the structure of CDR3 and amino acid sequences.
Also, based on the structural classification of VHHs, we designed a humanized synthetic VHH library calledPharmaLogical® Library. Importantly, this library retains the structural features contributing to the paratope formation on the camelid VHHs.
- Humanized FRs
Because VHH is a camelid antibody and VHH may have immunogenicity for humans, VHH may cause allergic reactions when it is dosed into the human body. Therefore, humanization of VHH structure is one of the most important processes for drug discovery with VHH. PharmaLogical® Library is designed based on the sequence of DP-47, widely known as a humanized reference, and VHHs on the market or in clinical trials. Thus, the VHH candidates which are selected in the screening process are not required a humanized process.
- Huge diversity of library-10¹³⁻¹⁴
The three CDRs (Complementarity Determining Regions) that form the antigen recognition site are designed based on the structural property information obtained from the alpaca-derived VHH. CDR3 is highly related to antigen binding. Therefore, we randomized the amino acid sequences of CDR3s, and it allows large diversity of VHHs. Also, our cDNA display method can handle this huge diversity ≧10¹³⁻¹⁴ without reduction of diversity size.
- Minimize heterogeneity-Design to minimize the frequency on occurrence of amino acids that cause heterogeneity in formulation
Removed amino acids related to aggregation in CMC in advance. Expect to significantly reduce the cost and period for downstream processes.
- Design of PharmaLogical® Library
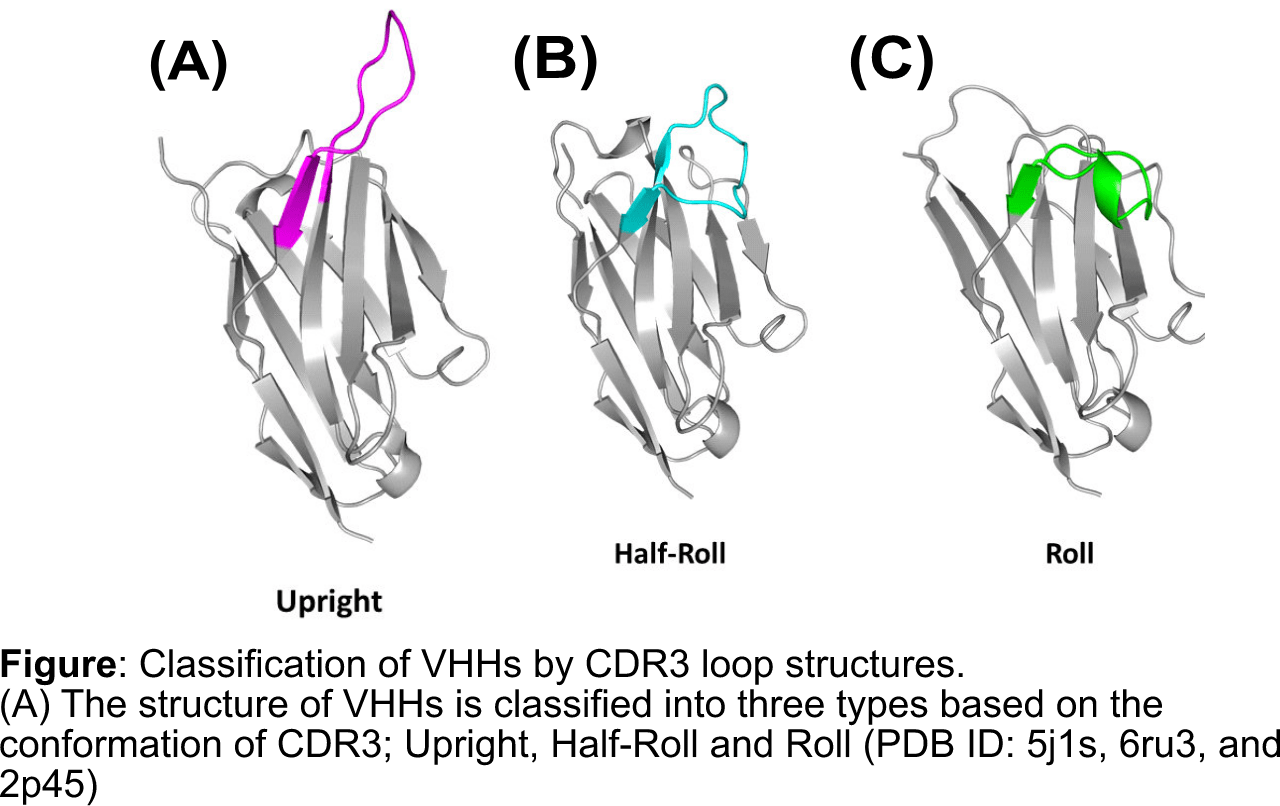
Based on research and these data, we designed PharmaLogical® Library
We designed two sub-libraries for PharmaLogical® Library that VHHs have either Upright and Roll CDR3, and these libraries include unique amino acids which codes for a unique length of CDR and frameworks.Reference:Murakami, T., Kumachi, S., Matsunaga, Y., Sato, M., Wakabayashi-Nakao, K., Masaki, H., Yonehara, R., Motohashi, M., Nemoto, N., & Tsuchiya, M. (2022). Construction ofa Humanized Artificial VHH Library Reproducing Structural Features of Camelid VHHs for Therapeutics. Antibodies (Basel), 11(1). https://doi.org/10.3390/antib11010010
- EME’s cyclic peptide library
In EME, we also own a cyclic peptide library.
- The Advantages of EME’s cyclic peptide library
- Formation of cyclic peptides with the chemical linker (increases stability)
- The hydrophobic/hydrophilic amino acids rich library
- 8 to 13 amino acids (equally randomized)
Reference:AnzaiH, Terai T, Wakabayashi-Nakao K, et al. Interleukin-17A Peptide Aptamers with an Unexpected Binding Moiety Selected by cDNA Display under Heterogenous Conditions. ACS Medicinal Chemistry Letters. 2021 Sep;12(9):1427-1434. DOI: 10.1021/acsmedchemlett.1c00217. PMID: 34531951.